德国picotweezers品牌,高分辨率三维单细胞单分子力学捕获分析光镊系统(3D Force Sensitive Optical Tweezers) PicoTweezers是一种结合了光镊技术及微视图像计算集合的分子生物力学分析系统。PicoTweezers作为一个独立的系统,可以与蔡司Axiovert、AxioA1或D1显微镜联合使用。PicoTweezers配备了功率为1W或5W的红外光纤激光器,可达激光陷阱捕获力范围是400pN——2nN。PicoTweezers的3D-压电平台可以实现x轴和y轴为200μn;m的分辨率,在z轴方向可以实现20μn;m的分辨率。独特的视频分析系统(Video-analysis)可以达到至少2.5纳米的横向和轴向分辨率,其图像拍摄速率为200帧/秒,X、Y、Z互相成像速度为400赫兹,可对生物大分子进行0.1PN作用力分辨率的实时分析。
|
英文介绍Introduction
Our Optical Tweezers System Provides:
Quantitative 3D Force Measurements with 0.1 pN Resolution
Achievable Trapping Force of 400 pN with 1 W Fiber Laser
Manipulation of Trapped Objects with Nanometer Precision
Compact and Ultrastable Modular System
Programmable LabView? Software Interface
Easy-to-use Force Calibration without Detector Alignment
Scope of Applications:
from Single Molecules to Living Cells
from Polymer Elasticity to Microfluidics
from Molecular Interactions to Nanopores
Optical tweezers are used to trap and actively manipulate microscopic objects. They also offer a vast area of applications by measuring forces applied to trapped objects.
The Optical TrapMicroscopic objects - like inpidual nano- or microparticles, cells, cell compartments, single or clustered molecules - can be trapped securely inside the center of a strongly focused laser beam.
When an external forces is acting on the trapped object, it deflects from the center of the trap as the deflection x depends linearly on trap stiffness k and force F.
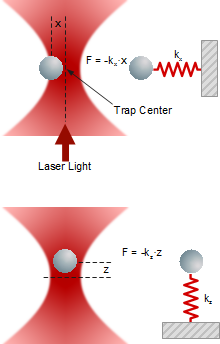 Lateral and axial forces acting on a trapped particleForces
Lateral and axial forces acting on a trapped particleForcesA trapped particle experiences various external forces. Atoms or molecules of the surrounding medium induce Brownian motion in all three dimensions, depending on temperature, viscosity and the presence of obstacles in the vicinity. Macroscopic fluid movements cause a drag force. Electric fields and bulk or surface charges may generate electrophoretic or electroosmotic forces.
Particularly, single molecules can induce forces of broad variety and magnitude while bound to the trapped object. On the other hand, the application of a force generated by an optical trap to a single molecule will gain vast insight into the molecular structure and elasticity, binding properties and kinetics.
Deflection is the EssenceGenerating and metering various forces requires a reliable force measurement capability in all three dimensions to allow for a maximum degree of experimental freedom and versatility. Therefore, force detection is accomplished by precisely measuring the deflection of the trapped particle in each direction.
The PicoTweezers system utilizes a sophisticated and easy-to-use video analysis (right image below) for particle tracking, detection and force measurements. It offers the largest field of application since it clears common calibration difficulties, system instabilities, as well as experimental and spatial restrictions.
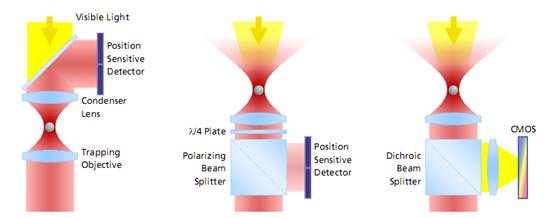
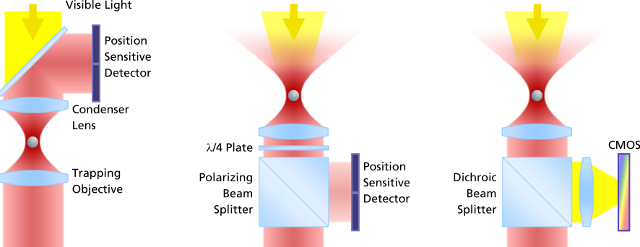 The evolution of force measurement. Left: Laser light passing trough the trapped particle (forward scattered light) is collected and projected onto a detector sensing the particle’s deflection. The condenser in close proximity to the trapping objective needs to be precisely adjusted. It is susceptible against drift and misalignment and limits the experimental space. Center: Backscattered light from the particle is collected by the trapping objective, separated from the incident laser light and projected onto the detector. This extremely robust setup allows high experimental freedom – the same applies for video-based detection method in the right image, where no detector alignment is required, too. In addition, a high persity of trapped particles can be video-analyzed and measured.Video Detection and Analysis
The evolution of force measurement. Left: Laser light passing trough the trapped particle (forward scattered light) is collected and projected onto a detector sensing the particle’s deflection. The condenser in close proximity to the trapping objective needs to be precisely adjusted. It is susceptible against drift and misalignment and limits the experimental space. Center: Backscattered light from the particle is collected by the trapping objective, separated from the incident laser light and projected onto the detector. This extremely robust setup allows high experimental freedom – the same applies for video-based detection method in the right image, where no detector alignment is required, too. In addition, a high persity of trapped particles can be video-analyzed and measured.Video Detection and AnalysisVideo-based force detection is easy to calibrate and provides an alignment-free and unsusceptible method for all force measurements in three dimensions. It is embedded into the LabView? platform.
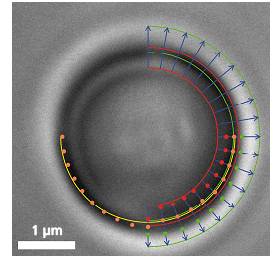
The Principle Video frame of a trapped microbead with various overlaid detection lines
Video frame of a trapped microbead with various overlaid detection linesIn addition to a video camera imaging the surrounding area of the optical trap, a second high-speed CMOS camera simultaneously surveys the magnified image of the trapped particle. The software searches for specific edges (as shown in the upper right quadrant of the image) in each frame and in real-time, determines gradients (lower right quadrant) and fits a circle (lower left quadrant) which correlates to the apparent particle diameter.
If an external axial force is acting on the particle, its apparent diameter changes, which the software translates into a z-force. Lateral forces only shift the center of the particle. These lateral deflections in the order of nanometers are then translated into x- and y-forces.
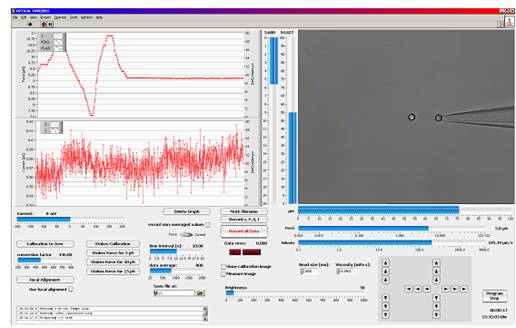
Easy and Reliable Force Calibration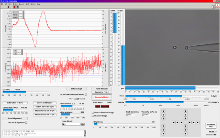 LabView? based trapping, calibration and measurement software
LabView? based trapping, calibration and measurement softwareForce calibration in three dimensions is conducted by moving the surrounding medium via the piezo stage using Stokes’ drag force law.
For specific applications, video-based force detection utilizes Allan Variance analysis and calibration. Here, smallest particle fluctuations are recorded and analyzed without the need of applying any frictional force. Both Stokes’ method and Allan Variance do not require the determination of the trap stiffness k, though the video-analysis software can calculate it if desired.
Benefits of Video-Based Force DetectionThere is no need of detector alignment or adjustment in the beginning or during experimentation because the CMOS camera providing data for video detection and analysis is integrated into the optical pathway between laser and optical trap. Video detection is unsusceptible to disturbing particles that occasionally may be trapped together with the measured object. Since the diameter of the trapped object is permanently monitored, further particles of interest can be trapped and compared with previous ones. Specifically tailored Allan Variance for video analysis is a powerful calibration tool for experiments that take place in an environment that prevents other calibration or analysis methods. When trapping particles close to interfaces (bottom or ceiling of sample chamber, artificial or biological membranes, etc.), video analysis delivers an interference-free force signal.
Detection TandemOptionally, PicoTweezers can be equipped with additional backscattered light detection capability for simultaneous measurements or as stand-alone method, if experiments need to be conducted in absence of light or if particle fluctuations must be analyzed with highest sample rate in the kHz range.
Applications — Single Molecules and Polymer ElasticityThe elastic behavior of a single DNA-strand in absence or in presence of binding ligands can be reliably measured. Theoretical polymer models that are fitted to the results will deliver parameters, which characterize the polymer elasticity.
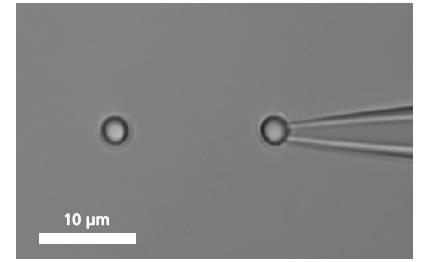
Grabbing a Single Molecule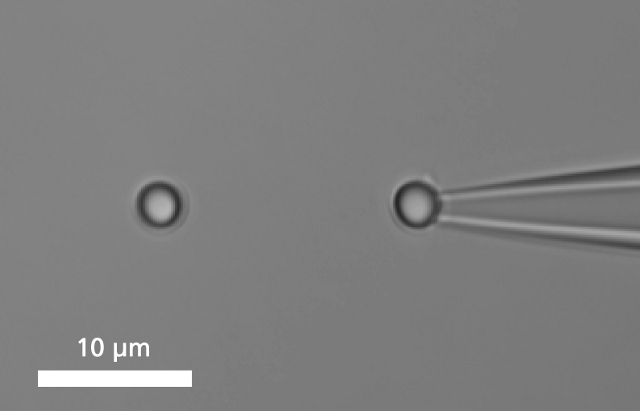 A single DNA is immobilized between two microbeads.
A single DNA is immobilized between two microbeads.To bind a single DNA-strand between two coated microbeads, it has to be properly functionalized on both ends. Thus, it can be immobilized between two beads, of which one is optically trapped and the other is held on the tip of a micropipette. Increasing the distance between the beads by moving the piezo stage induces a controlled mechanical tension to the DNA.
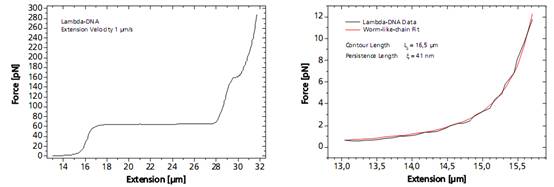
As a response, the force-extension curve of the molecule exhibit characteristic mechanical properties, such as an entropic elasticity, an overstretching plateau and a melting transition region.
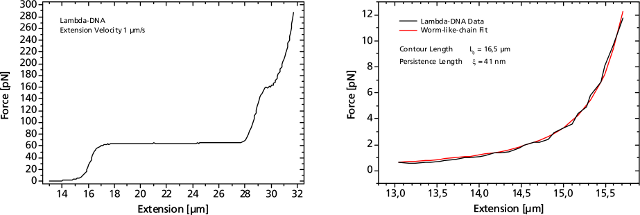 Left: Force response of a single 48502 base pair long DNA molecule of bacteriophage lambda. In the force range up to 10 pN the entropic regime determines the elastic behavior of the molecule, whereas around 65 pN the characteristic overstretching transition occurs. The nature of this phenomena remains controversial, as well as for a less pronounced transition at 160 pN. Right: Fitting the Worm-like-chain model to the entropic regime yields two intrinsic elasticity parameters. For example, the persistence length strongly depends on salt concentration and on the presence of DNA-binding ligands.DNA as Sensor for Foreign Molecules
Left: Force response of a single 48502 base pair long DNA molecule of bacteriophage lambda. In the force range up to 10 pN the entropic regime determines the elastic behavior of the molecule, whereas around 65 pN the characteristic overstretching transition occurs. The nature of this phenomena remains controversial, as well as for a less pronounced transition at 160 pN. Right: Fitting the Worm-like-chain model to the entropic regime yields two intrinsic elasticity parameters. For example, the persistence length strongly depends on salt concentration and on the presence of DNA-binding ligands.DNA as Sensor for Foreign MoleculesThe DNA strand can serve as a host for a variety of different molecules, such as small intercalators, groove-binders, proteins, enzymes or molecular motors.
The binding event of a single or a multitude of ligands can change the elastic response more or less significantly. As an example, the force curve of a DNA is shown in presence of the antibiotic distamycin-A that attaches to the minor groove of the DNA strand while stabilizing it and helping to resist the overstretching. On the other hand, diazoniapentaphene as an intercalator increases both contour and persistence length and renders the overstretching plateau to disappear.
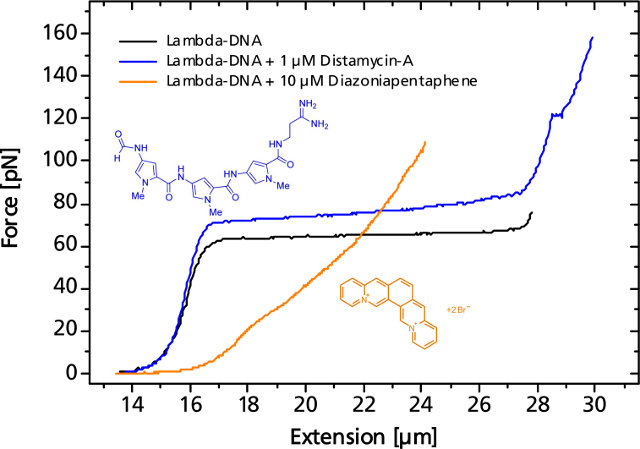 Small or large variations in the elastic response of a DNA-molecule in the presence of binding ligands can be measured.Applications — Translocation through Nanopores
Small or large variations in the elastic response of a DNA-molecule in the presence of binding ligands can be measured.Applications — Translocation through NanoporesNanopores play a major role in biology and they rapidly evolved into a new and promising technique in single-molecule detection. The controlled threading of a single DNA molecule or a DNA-protein complex into a nanopore allows investigation of the translocation dynamics and a localization of the bound protein.
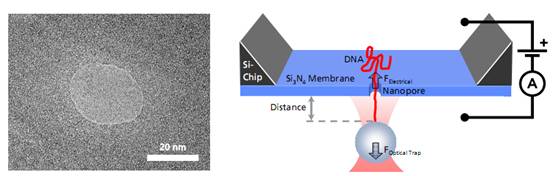
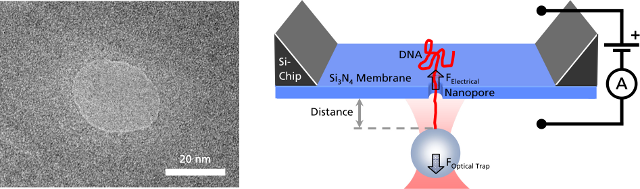 Left: TEM image of a solid-state nanopore drilled with a focused ion beam machine into a Si3N4membrane that serves as model system to study single molecule translocations. Right: Experimental setup of a DNA translocation measured with optical tweezers. When applying a voltage across the membrane, a single DNA molecule immobilized on a trapped microbead translocates through the pore. The electrostatic force acting on the molecule and the distance between bead and nanopore can be precisely measured.Translocating a Single DNA Strand
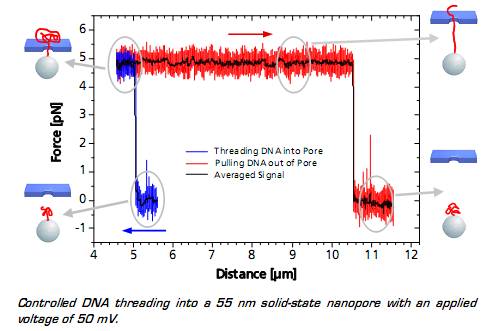
Left: TEM image of a solid-state nanopore drilled with a focused ion beam machine into a Si3N4membrane that serves as model system to study single molecule translocations. Right: Experimental setup of a DNA translocation measured with optical tweezers. When applying a voltage across the membrane, a single DNA molecule immobilized on a trapped microbead translocates through the pore. The electrostatic force acting on the molecule and the distance between bead and nanopore can be precisely measured.Translocating a Single DNA StrandWhen the DNA on the trapped bead approaches the nanopore (to a distance of 5 μm) it is immediately threaded into the pore by electrostatic forces acting on the negatively charged DNA backbone. This effect can be monitored as an abrupt step of the force signal to a certain value, which remains constant even when retracting the bead. The measured force depends on the applied voltage, as well as on the diameter of the nanopore.
When the entire DNA strand with an end-end-distance of 10.5 μm is pulled out of the pore, the force drops back to zero.
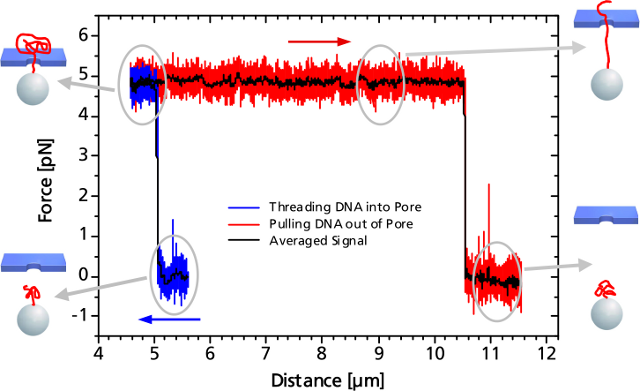 Controlled DNA threading into a 55 nm solid-state nanopore with an applied voltage of 50 mV.Single DNA-Bound Protein
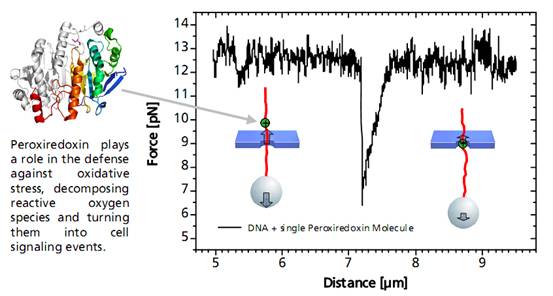
Controlled DNA threading into a 55 nm solid-state nanopore with an applied voltage of 50 mV.Single DNA-Bound ProteinA distinct asymmetric force signal occurs when a single peroxiredoxin molecule bound to the DNA stand is actively pulled through the pore. This effect serves as a label-free localization of the protein binding site.
It can be understand as the result of an effective positive charge of the protein counteracting the negative DNA backbone charge and reducing the electrostatic force.
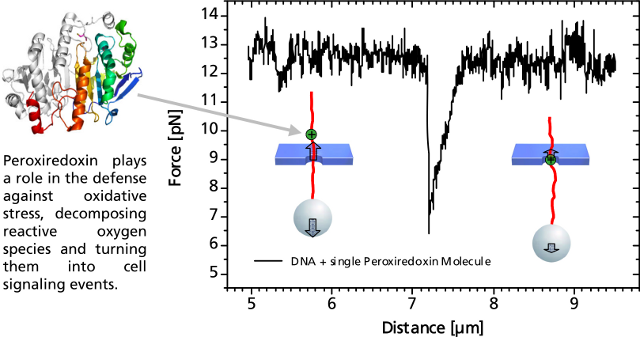 Characteristic force signal of a single peroxiredoxin molecule bound to a DNA strand when both are translocated through a 35 nm nanopore.We Design and Build Your Optical Tweezers.
Characteristic force signal of a single peroxiredoxin molecule bound to a DNA strand when both are translocated through a 35 nm nanopore.We Design and Build Your Optical Tweezers.PicoTweezers is a stand-alone system, that can also be customized to your Zeiss Axiovert, Axio Observer A1 or D1. It will be equipped with a 1 W or 5 W IR fiber laser for highest spatial trap stability yielding a trapping force of at least 400 pN or 2 nN, respectively.
The 3D-piezo stage enables nanometer resolution in a range of 200 μm in x and y, as well as 20 μm in z-direction. Video-analysis can achieve a lateral and axial resolution of at least 2.5 nm, which results in a force resolution of 0.1 pN with a frame rate of 200 and 400 Hz in z- and x,y-direction, respectively
ReferencesS. Knust, A. Spiering, H. Vieker, A. Beyer, A. G?lzh?user, K. T?nsing, A. Sischka and D. Anselmetti
Video-Based and Interference-Free Axial Force Detection and Analysis for Optical Tweezers
Review of Scientific Instruments, 83, 103704 (2012)A. Spiering, S. Getfert, A. Sischka, P. Reimann and D. Anselmetti
Nanopore Translocation Dynamics of a Single DNA-Bound Protein
Nano Letters, 11, 2978 (2011)A. Sischka, A. Spiering, M. Khaksar, M. Laxa, J. K?nig, K.J. Dietz and D. Anselmetti
Dynamic Translocation of Ligand-Complexed DNA Through Solid-State Nanopores with Optical Tweezers
Journal of Physics - Condensed Matter, 22, 454121 (2010)C. Kleimann, A. Sischka, A. Spiering, K. T?nsing, N. Sewald, U. Diedrichsen and D. Anselmetti
Binding Kinetics of Bisintercalator Triostin A with Optical Tweezers Force Mechanics
Biophysical Journal, 97, 2780 (2009)D. Pla, A. Sischka, F. Albericio, M. Alvarez, X. Fernandez-Busquets and D. Anselmetti
Optical-Tweezers Study of Topoisomerase Inhibition
Small, 5, 1269 (2009)A. Sischka, C. Kleimann, W. Hachmann, M.M. Sch?fer, I. Seuffert, K. T?nsing and D. Anselmetti
Single Beam Optical Tweezers Setup with Backscattered Light Detection for Three-Dimensional Measurements on DNA and Nanopores
Review of Scientific Instruments, 79, 063702 (2008)D. Anselmetti, N. Hansmeier, J. Kalinowski, J. Martini, T. Merkle, R. Palmisano, R. Ros, K. Schmied, A. Sischka and K. T?nsing
Analysis of Subcellular Surface Structure, Function and Dynamics
Analytical and Bioanalytical Chemistry, 387, 83 (2007)A. Sischka, K. T?nsing, R. Eckel, S.D. Wilking, N. Sewald, R. Ros and D. Anselmetti
Molecular Mechanisms and Kinetics between DNA and DNA Binding Ligands
Biophysical Journal, 88, 404 (2005)